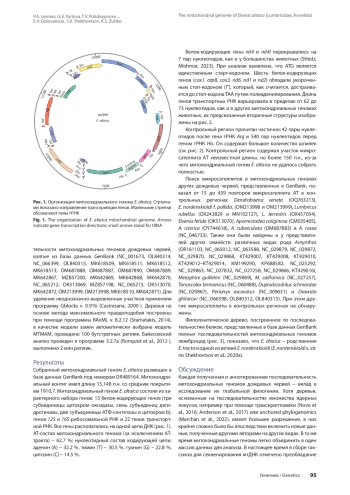
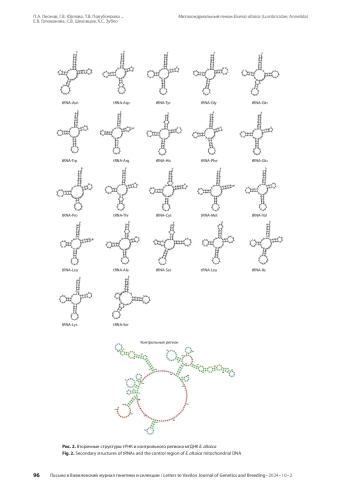
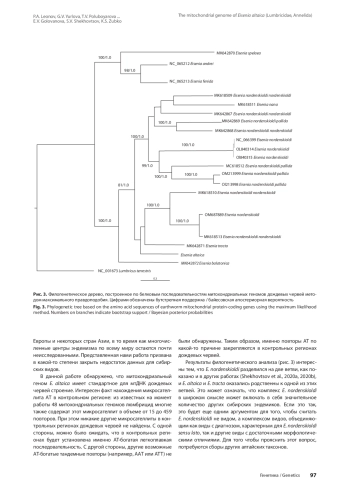
1. Всеволодова-Перель Т.С. Дождевые черви России: кадастр и определитель. М.: Наука, 1997.
Vsevolodova-Perel T.S. The Earthworms of the Russian Fauna: Cadaster and Key. Moscow: Nauka Publ., 1997 (in Russian).
2. Перель Т.С. Дождевые черви в почвах лесов Северо-Западного Кавказа. В: Влияние животных на продуктивность лесных биогеоценозов. М.: Наука, 1966;146-165.
Perel T.S. Earthworms in forest soils of the Northwestern Caucasus. In: The Impact of Animals on the Productivity of Forest Cenoses. Moscow: Nauka Publ., 1966;146-165 (in Russian).
3. Шеховцов С.В., Базарова Н.Э., Берман Д.И., Булахова Н.А., Голованова Е.В., Коняев С.В., Кругова Т.М., Любечанский И.И., Пельтек С.Е. ДНК-штрихкодирование: сколько видов дождевых червей живет на юге Западной Сибири? Вавиловский журнал генетики и селекции. 2016;20(1):125-130. DOI: 10.18699/VJ15.110 EDN: WORKTR
Shekhovtsov S.V., Bazarova N.E., Berman D.I., Bulakhova N.A., Golovanova E.V., Konyaev S.V., Krogova T.M., Lyubechanskii I.I., Peltek S.E. DNA barcoding: How many earthworm species are there in the south of West Siberia? Russian Journal of Genetics: Applied Research. 2017;7(1):57-62. DOI: 10.1134/S2079059717010130 EDN: YVGAET
4. Шеховцов С.В., Рапопорт И.Б., Полубоярова Т.В., Гераськина А.П., Голованова Е.В., Пельтек С.Е. Морфотипы и генетическая изменчивость Dendrobaena schmidti (Lumbricidae, Annelida). Вавиловский журнал генетики и селекции. 2020;24(1):48-54. DOI: 10.18699/VJ20.594 EDN: IELWGX
Shekhovtsov S.V., Rapoport I.B., Poluboyarova T.V., Geraskina A.P., Golovanova E.V., Peltek S.E. Morphotypes and genetic diversity of Dendrobaena schmidti (Lumbricidae, Annelida). Vavilovskii Zhurnal Genetiki i Selektsii = Vavilov Journal of Genetics and Breeding. 2020;24(1):48-54. 10.18699/VJ20.594 (in Russian). DOI: 10.18699/VJ20.594(inRussian) EDN: IELWGX
5. Anderson F.E., Williams B.W., Horn K.M., Erséus C., Halanych K.M., Santos S.R., James S.W. Phylogenomic analyses of Crassiclitellata support major Northern and Southern Hemisphere clades and a Pangaean origin for earthworms. BMC Evol. Biol. 2017;17(1):123. DOI: 10.1186/s12862-017-0973-4 EDN: NPBDGG
6. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573-580. DOI: 10.1093/nar/27.2.573 EDN: IUXIWX
7. Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69(2):313-319. DOI: 10.1016/j.ympev.2012.08.023
8. Briones M.J.I., Morán P., Posada D. Are the sexual, somatic and genetic characters enough to solve nomenclatural problems in lumbricid taxonomy? Soil Biol. Biochem. 2009;41(11):2257-2271. DOI: 10.1016/j.soilbio.2009.07.008 EDN: KJIDTF
9. Eisen G. On the Oligochaeta collected during the Swedish expeditions to the arctic regions in the years 1870, 1875 and 1876. Kongl. Sv. Vetensk. Akad. Handl. 1879;15:1-49.
10. Gendron P., Lemieux S., Major F. Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol. 2001;308(5):919-936. DOI: 10.1006/jmbi.2001.4626 EDN: KFNLPF
11. Jin J.J., Yu W.B., Yang J.B., Song Y., DePamphilis C.W., Yi T.S., Li D.Z. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. DOI: 10.1186/s13059-020-02154-5 EDN: FHEVBX
12. Marchán D.F., James S.W., Lemmon A.R., Lemmon E.M., Novo M., Domínguez J., Cosín D.J.D., Trigo D. A strong backbone for an invertebrate group: anchored phylogenomics improves the resolution of genuslevel relationships within the Lumbricidae (Annelida, Crassiclitellata). Org. Divers. Evol. 2022;22(4):915-924. DOI: 10.1007/s13127-022-00570-y EDN: LWBWNZ
13. Novo M., Fernández R., Andrade S.C.S., Marchán D.F., Cunha L., Díaz Cosín D.J. Phylogenomic analyses of a Mediterranean earthworm family (Annelida: Hormogastridae). Mol. Phylogenet. Evol. 2016;94(Pt. B):473-478. DOI: 10.1016/j.ympev.2015.10.026
14. Porebski S., Bailey L.G., Baum B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997;15(1):8-15. DOI: 10.1007/BF02772108 EDN: XSNALF
15. Shtolz N., Mishmar D. The metazoan landscape of mitochondrial DNA gene order and content is shaped by selection and affects mitochondrial transcription. Commun. Biol. 2023;6(1):93. DOI: 10.1038/s42003-023-04471-4 EDN: XFQCDG
16. Shekhovtsov S.V., Ershov N.I., Vasiliev G.V., Peltek S.E. Transcriptomic analysis confirms differences among nuclear genomes of cryptic earthworm lineages living in sympatry. BMC Evol. Biol. 2019;19(1):13-22. DOI: 10.1186/s12862-019-1370-y EDN: WUSRPU
17. Shekhovtsov S.V., Shipova A.A., Poluboyarova T.V., Vasiliev G.V., Golovanova E.V., Geraskina A.P., Bulakhova N.A., Szederjesi T., Peltek S.E. Species delimitation of the Eisenia nordenskioldi complex (Oligochaeta, Lumbricidae) using transcriptomic data. Front. Genet. 2020a;11:598196. DOI: 10.3389/fgene.2020.598196 EDN: ZHHHDB
18. Shekhovtsov S.V., Golovanova E.V., Ershov N.I., Poluboyarova T.V., Berman D.I., Bulakhova N.A., Szederjesi T., Peltek S.E. Phylogeny of the Eisenia nordenskioldi complex based on mitochondrial genomes. Eur. J. Soil Biol. 2020b;96:103137. 10.1016/j.ejsobi. 2019.103137. DOI: 10.1016/j.ejsobi.2019.103137 EDN: GHYLRO
19. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312-1313. DOI: 10.1093/bioinformatics/btu033