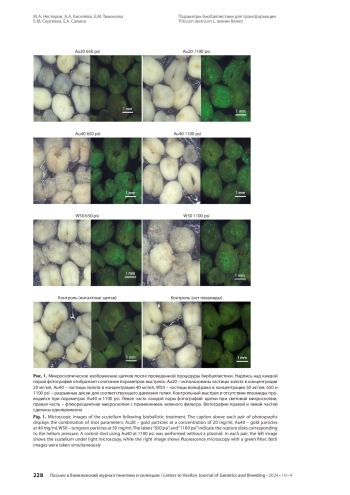
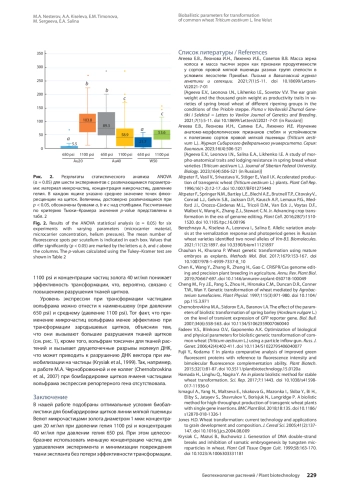
Для улучшения хозяйственно ценных признаков мягкой пшеницы ( Triticum aestivum L.) весьма перспективно геномное редактирование, а биобаллистический метод - один из наиболее распространенных способов доставки генетических конструкций. Бóльшая часть опубликованных работ по редактированию мягкой пшеницы выполнена с использованием нескольких модельных сортов. Показано, что эффективность трансформации - генотип-специфичный показатель, поэтому подбор условий для успешной трансформации немодельных генотипов является актуальной задачей. В работе проведено сравнение эффективности биобаллистики для трансформации зародышевых щитков мягкой пшеницы линии Велют при варьировании следующих параметров: материал и концентрация микрочастиц (20, 40 мг/мл для золотых микрочастиц и 50 мг/мл для вольфрамовых), давление гелия (650 и 1100 psi). Эффективность биобаллистики оценивали по среднему числу клеток, экспрессирующих репортерный ген белка eGFP, на эксплант. Результаты показали, что при использовании частиц вольфрама как при 650 psi, так и 1100 psi, а также частиц золота при 1100 psi и 40 мг/мл эффективность трансформации снижается из-за усиления повреждения тканей щитков. Наибольшая эффективность бомбардировки отмечена для микрочастиц золота при следующих сочетаниях параметров: концентрация частиц 20 мг/мл и давление 1100 psi либо 40 мг/мл и давление 650 psi.
Идентификаторы и классификаторы
Для двудольных растений успешно используют перенос генетического материала с помощью агробактерий, метод не требует дорогостоящего оборудования и дает стабильные результаты. Однако для растений из класса однодольных, в частности семейства злаковые (Poaceae Barnh.), применение агробактериальной трансформации ограничено по ряду причин: в целом менее эффективная регенерация in vitro, уровень которой сильно зависит от генотипа; отсутствие природной восприимчивости однодольных к агробактериям. Успешные результаты агробактериальной трансформации показаны для отдельных сортов пшеницы и ячменя (Wan, Lemaux, 1994; Cheng et al., 1997).
Список литературы
1. Агеева Е.В., Леонова И.Н., Лихенко И.Е., Советов В.В. Масса зерна колоса и масса тысячи зерен как признаки продуктивности у сортов яровой мягкой пшеницы разных групп спелости в условиях лесостепи Приобья. Письма в Вавиловский журнал генетики и селекции. 2021;7(1):5-11. DOI: 10.18699/Letters-VJ2021-7-01 EDN: XLSVJV
Ageeva E.V., Leonova I.N., Likhenko I.E., Sovetov V.V. The ear grain weight and the thousand grain weight as productivity traits in varieties of spring bread wheat of different ripening groups in the conditions of the Priob’e steppe. Pisma v Vavilovskii Zhurnal Genetiki i Selektsii = Letters to Vavilov Journal of Genetics and Breeding. 2021;7(1):5-11. 10.18699/LettersVJ2021-7-01 (in Russian). DOI: 10.18699/LettersVJ2021-7-01(inRussian) EDN: XLSVJV
2. Агеева Е.В., Леонова И.Н., Салина Е.А., Лихенко И.Е. Изучение анатомо-морфологических признаков стебля и устойчивости к полеганию сортов яровой мягкой пшеницы (Triticum aestivum L.). Журнал Сибирского федерального университета. Серия: Биология. 2023;16(4):506-521. EDN: TCSUQF
Ageeva E.V., Leonova I.N., Salina E.A., Likhenko I.E. A study of morpho-anatomical traits and lodging resistance in spring bread wheat varieties (Triticum aestivum L.). Journal of Siberian Federal University. Biology. 2023;16(4):506-521 (in Russian). EDN: TCSUQF
3. Altpeter F., Vasil V., Srivastava V., Stöger E., Vasil I.K. Accelerated production of transgenic wheat (Triticum aestivum L.) plants. Plant Cell Rep. 1996;16(1-2):12-17. DOI: 10.1007/BF01275440
4. Altpeter F., Springer N.M., Bartley L.E., Blechl A.E., Brutnell T.P., Citovsky V., Conrad L.J., Gelvin S.B., Jackson D.P., Kausch A.P., Lemaux P.G., Medford J.I., Orozco-Cárdenas M.L., Tricoli D.M., Van Eck J., Voytas D.F., Walbot V., Wang K., Zhang Z.J., Stewart C.N. Jr. Advancing crop transformation in the era of genome editing. Plant Cell. 2016;28(7):1510-1520. DOI: 10.1105/tpc.16.00196
5. Berezhnaya A., Kiseleva A., Leonova I., Salina E. Allelic variation analysis at the vernalization response and photoperiod genes in Russian wheat varieties identified two novel alleles of Vrn-B3. Biomolecules. 2021;11(12):1897. DOI: 10.3390/biom11121897 EDN: BRPNUS
6. Chauhan H., Khurana P. Wheat genetic transformation using mature embryos as explants. Methods Mol. Biol. 2017;1679:153-167. DOI: 10.1007/978-1-4939-7337-8_10 EDN: QGKTAD
7. Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667-697. DOI: 10.1146/annurev-arplant-050718-100049 EDN: GMCCLX
8. Cheng M., Fry J.E., Pang S., Zhou H., Hironaka C.M., Duncan D.R., Conner T.W., Wan Y. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 1997;115(3):971-980. DOI: 10.1104/pp.115.3.971
9. Chernobrovkina M.A., Sidorov E.A., Baranov I.A. The effect of the parameters of biolistic transformation of spring barley (Hordeum vulgare L.) on the level of transient expression of GFP reporter gene. Biol. Bull. 2007;34(6):558-563. DOI: 10.1134/S1062359007060040 EDN: LKTVDZ
10. Fadeev V.S., Blinkova O.V., Gaponenko A.K. Optimization of biological and physical parameters for biolistic genetic transformation of common wheat (Triticum aestivum L.) using a particle inflow gun. Russ. J. Genet. 2006;42(4):402-411. DOI: 10.1134/S1022795406040077 EDN: LJVROR
11. Fujii Y., Kodama Y. In planta comparative analysis of improved green fluorescent proteins with reference to fluorescence intensity and bimolecular fluorescence complementation ability. Plant Biotech. 2015;32(1):81-87. DOI: 10.5511/plantbiotechnology.15.0120a EDN: UKDEAI
12. Hamada H., Linghu Q., Nagira Y. An in planta biolistic method for stable wheat transformation. Sci. Rep. 2017;7:11443. DOI: 10.1038/s41598-017-11936-0 EDN: NBXOEX
13. Ismagul A., Yang N., Maltseva E., Iskakova G., Mazonka I., Skiba Y., Bi H., Eliby S., Jatayev S., Shavrukov Y., Borisjuk N., Langridge P. A biolistic method for high-throughput production of transgenic wheat plants with single gene insertions. BMC Plant Biol. 2018;18:135. DOI: 10.1186/s12870-018-1326-1 EDN: YBMZFZ
14. Jones H.D. Wheat transformation: current technology and applications to grain development and composition. J. Cereal Sci. 2005;41(2):137-147. DOI: 10.1016/j.jcs.2004.08.009
15. Krysiak C., Mazuś B., Buchowicz J. Generation of DNA double-strand breaks and inhibition of somatic embryogenesis by tungsten microparticles in wheat. Plant Cell Tissue Organ Cult. 1999;58:163-170. :1006303331181. DOI: 10.1023/A EDN: AGMOTD
16. Lonsdale D., Onde S., Cuming A. Transient expression of exogenous DNA in intact, viable wheat embryos following particle bombardment. J. Exp. Bot. 1990;41(9):1161-1165. DOI: 10.1093/jxb/41.9.1161 EDN: IRHZMZ
17. Matveev A.V., Nartova A.V., Sankova N.N., Okunev A.G. DLgram cloud service for deep-learning analysis of microscopy images. Microsc. Res. Tech. 2024;87(5):991-998. DOI: 10.1002/jemt.24480 EDN: KLTGLG
18. Miroshnichenko D., Filippov M., Dolgov S. Genetic transformation of Russian wheat cultivars. Biotechnol. Biotechnol. Equip. 2007;21(4):399-402. DOI: 10.1080/13102818.2007.10817482 EDN: LKHBMD
19. Miroshnichenko D.N., Poroshin G.N., Dolgov S.V. Genetic transformation of wheat using mature seed tissues. Appl. Biochem. Microbiol. 2011;47:767-775. DOI: 10.1134/S0003683811080096 EDN: PEFDFH
20. Miroshnichenko D., Ashin D., Pushin A., Dolgov S. Genetic transformation of einkorn (Triticum monococcum L. ssp. monococcum L.), a diploid cultivated wheat species. BMC Biotechnol. 2018;18:68. DOI: 10.1186/s12896-018-0477-3 EDN: TLRIGV
21. Miroshnichenko D.N., Klementyeva A.A., Salina E.A., Dolgov S.V. Evaluation of in vitro plant regeneration efficiency in Siberian wheat cultivars. In: Current Challenges in Plant Genetics, Genomics, Bioinformatics, and Biotechnology. Proceedings of the Fifth International Scientific Conference PlantGen-2019. Novosibirsk, 2019;126-128. DOI: 10.18699/ICG-PlantGen2019-40 EDN: MIKPJC
22. Miroshnichenko D., Klementyeva A., Pushin A., Dolgov S. A competence of embryo-derived tissues of tetraploid cultivated wheat species Triticum dicoccum and Triticum timopheevii for efficient and stable transgenesis mediated by particle inflow gun. BMC Plant Biol. 2020;20(Suppl. 1):442. DOI: 10.1186/s12870-020-02580-4 EDN: GZRCYF
23. Pang S.Z., DeBoer D.L., Wan Y., Ye G., Layton J.G., Neher M.K., Armstrong C.L., Fry J.E., Hinchee M.A., Fromm M.E. An improved green fluorescent protein gene as a vital marker in plants. Plant Physiol. 1996;112(3):893-900. DOI: 10.1104/pp.112.3.893
24. PDS-1000/He Biolistic Particle Delivery System Instruction Manual. [https://www.biorad.com/sites/default/files/webroot/web/pdf/lsr/literature/10000070900.pdf].
25. Rasco-Gaunt S., Riley A., Barcelo P., Lazzeri P.A. Analysis of particle bombardment parameters to optimise DNA delivery into wheat tissues. Plant Cell Rep. 1999;19:118-127. DOI: 10.1007/s002990050721 EDN: AVNGKJ
26. Rasco-Gaunt S., Riley A., Cannell M., Barcelo P., Lazzeri P.A. Procedures allowing the transformation of a range of European elite wheat (Triticum aestivum L.) varieties via particle bombardment. J. Exp. Bot. 2001;52(357):865-874. DOI: 10.1093/jexbot/52.357.865 EDN: HUUNEY
27. Russell B.C., Torralba A., Murphy K.P., Freeman W.T., LabelMe: a database and web-based tool for image annotation. Int. J. Comput. Vis. 2008;77(1-3):157-173. DOI: 10.1007/s11263-007-0090-8 EDN: NAYUNC
28. Sanford J.C., Smith F.D., Russell J.A. Optimizing the biolistic process for different biological applications. Methods Enzymol. 1993;217:483-509. DOI: 10.1016/0076-6879(93)17086-k EDN: XZEIWG
29. Souza C., Eduardo D., Fettig S., Ziegler P., Beck E. Transformation of an Argentine spring wheat genotype: optimization of the protocols for particle bombardment of excised immature embryos and rapid isolation of transgenic plants. BAG. J. Basic Appl. Genet. 2015;26(1):18-37. EDN: USARZJ
30. Sparks C.A., Doherty A. Genetic transformation of common wheat (Triticum aestivum L.) using biolistics. Methods Mol Biol. 2020;2124:229-250. DOI: 10.1007/978-1-0716-0356-7_12
31. Tian B., Navia-Urrutia M., Chen Y., Brungardt J., Trick H.N. Biolistic transformation of wheat. Methods Mol. Biol. 2018;1864:117-130. DOI: 10.1007/978-1-4939-8778-8_9
32. Vasil V., Castillo A., Fromm M., Vasil I.K. herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Nat. Biotechnol. 1992;10:667-674. DOI: 10.1038/nbt0692-667
33. Wada K. Labelme: Image Polygonal Annotation with Python [Computer software]. 2021. DOI: 10.5281/zenodo.5711226
34. Wan Y., Lemaux P.G. Generation of large numbers of independently transformed fertile barley plants. Plant Physiol. 1994;104(1):37-48. DOI: 10.1104/pp.104.1.37 EDN: NXUJXD
35. Wang Y., Zeng J., Su P., Zhao H., Li L., Xie X., Zhang Q., Wu Y., Wang R. Zhang Y., Yu B., Chen M., Wang Y., Yang G., He G., Chang J., Li Y. An established protocol for generating transgenic wheat for wheat functional genomics via particle bombardment. Front. Plant Sci. 2022;13:979540. DOI: 10.3389/fpls.2022.979540 EDN: NBIUWQ
36. Yao Q., Cong L., He G.Y., Chang J.L., Li K.X., Yang G.X. Optimization of wheat co-transformation procedure with gene cassettes resulted in an improvement in transformation frequency. Mol. Biol. Rep. 2007;34(1):61-67. DOI: 10.1007/s11033-006-9016-8 EDN: AGRGBN
37. Zhang K., Liu J., Zhang Y., Yang Z., Gao C. Biolistic genetic transformation of a wide range of Chinese elite wheat (Triticum aestivum L.) varieties. J. Genet. Genomics. 2015;42(1):39-42. DOI: 10.1016/j.jgg.2014.11.005
Выпуск
Другие статьи выпуска
Обратимое метилирование мРНК - модификация N6-метиладенозина (m6A) - оказывает влияние почти на все стадии ее метаболизма. Динамические и обратимые процессы регулируются «записывающими» m6A-метилтрансферазами, «стирающими» m6A-деметилазами и «считывающими» m6A-связывающими белками. Эти регуляторы распознают, добавляют или удаляют сайты, модифицированные m6A, соответствующим образом изменяя биологические процессы. m6A присутствует во многих мРНК, кодируемых генами, связанными с заболеваниями человека, в том числе онкологическими. Роль модификации m6A мРНК в возникновении опухоли и ее прогрессии связана главным образом с активацией экспрессии онкогенов и подавлением экспрессии генов опухолевой супрессии. В зависимости от уровня метилирования аденозина, экспрессии и активности соответствующих ферментов эта модификация мРНК может приводить как к активации, так и к торможению роста опухоли. Показано участие m6A совместно с другими эпигенетическими модификациями в регуляции возникновения, развития и прогрессии опухоли, в частности в ангиогенезе. Молекулярный механизм действия метилтрансферазы METTL3 является возможной мишенью для диагностики и лечения онкологических заболеваний, что важно для практической медицины. Об этом говорит влияние на рост опухоли ингибиторов METTL3 и ангиогенеза, показавших эффективность при некоторых типах опухолей.
Аденоассоциированные вирусы (AAV) прочно вошли в практику научных исследований в широком спектре областей, от молекулярной биологии до физиологии. Они прошли путь от открытия их как вирусов в 1965 г. до широко используемого молекулярно-биологического инструмента на сегодняшний день. Исследователей привлекает в них надежность, стабильная экспрессия трансгена и низкая иммуногенность. Часто AAV становятся привлекательным средством доставки для генотерапии. Все больше фармацевтических компаний запускают клинические испытания с использованием AAV в качестве доставки гено-терапии. В 2023 г. Food and Drug Administration (FDA - Управление по контролю качества пищевых продуктов и лекарственных средств, США) был одобрен препарат Roctavian для лечения гемофилии A на основе AAV. Прогресс в этой области навел нас на мысль о его концептуальном обобщении и написании настоящей работы. В статье приведен анализ последних молекулярно-биологических и биотехнологических решений для аденоассоциированной вирусной доставки, а также ее оптимизации на животных моделях и способах сделать ее более направленной. Рассмотрены особенности серотипов аденоассоциированных вирусов, особое внимание уделено их тропизмам к клеткам организма и генно-инженерным способам их изменения - направленной эволюции капсидов, использованию химерных капсидов, сшитых с рецепторами или одноцепочечными антителами альпак. Существенным недостатком AAV является ограниченность кассеты - лишь 4.7 кб генного материала. В обзоре описаны приемы увеличения переносимого генетического материала и осуществления трансдукции длинных по протяженности до 10 кб последовательностей кДНК; собрана информация по проводимым клиническим испытаниям, в которых задействованы AAV, а также охарактеризованы проблемы реализации доставки генов при применении AAV в терапии.
Описана математическая модель расщепления, основанная на фундаментальных свойствах нормального распределения. Предложены классификация расщеплений и их соотнесение с методикой исследования, ориентированной на преимущественное использование количественных (измеряемых) признаков. Описан алгоритм последовательного разделения би- и мультимодальных выборок на отдельные группы с применением свойства симметрии нормального распределения. Представлен метод балансировки групп, повышающий точность деления исходной выборки и унифицирующий подсчет количества объектов в группах. Продемонстрирована применимость описываемого метода к сложным распределениям различного вида, обеспечивающая определение формулы расщепления для выявленных групп. Приведены сведения о доступе к исполняемому модулю и исходным текстам специально разработанного инструментального средства.
В последние годы в список модельных организмов внесен свободноживущий плоский червь Macrostomum lignano, нашедший широкое применение в ряде областей научных изысканий. Его ключевая особенность, высокий потенциал к регенерации, предоставляет ему устойчивость к токсичным соединениям и онкогенам, высокую адаптивность к резким изменениям факторов окружающей среды, а также длительный срок жизни, граничащий с условным бессмертием. С другой стороны, особенности хромосомного состава генома M. lignano, выраженные в ряде геномных нестабильностей, вкупе с регенерацией, не переходящей в опухолевый генез, открывает широкие возможности для фундаментальных исследований противораковых терапий. Обзор посвящен разбору направлений биологических наук, где применяется или мог бы применяться M. lignano.
Статистика статьи
Статистика просмотров за 2025 - 2026 год.
Издательство
- Издательство
- НИИТПМ
- Регион
- Россия, Новосибирск
- Почтовый адрес
- 630089, г. Новосибирск, ул. Б. Богаткова, 175/1, Метро "Золотая нива", Автобус "Молодежная, Кошурникова"
- Юр. адрес
- 630090, г. Новосибирск, пр-т Академика Лаврентьева, 10
- ФИО
- Рагино Юлия Игоревна (Руководитель)
- Контактный телефон
- +7 (383) 3730981
- Сайт
- https://iimed.ru/